In abundant spare time 🕰️, yours truly has removed ❌ the non-linear 〰️ terms from the Pressure Poisson Equation (PPE) which is derived 🎡 by summing the divergence of momentum equations and then applying conservation of mass ⚖️. The non-linear terms create a problem in convergence. Therefore, inspired by the style of work of the "Skipper"🐧, yours truly removed the problem causing non-linear terms 😀. The derived PPE is mentioned by equation 1. The PPE used in the code yours truly has developed is mention in equation 2. Within equations 1 and 2, the u and v are components of velocity along x and y-axis. The pressure is represented by p and while, t represents the time.
∂2p/∂x2 + ∂2p/∂y2 = 1/∆t * (∂u/∂x + ∂v/∂y - (∂u/∂x)2 - (∂v/∂y)2 - 2*∂v/∂y*∂u/∂x) [1]
∂2p/∂x2 + ∂2p/∂y2 = 1/(2 * ∆t) * (∂u/∂x + ∂v/∂y) [2]
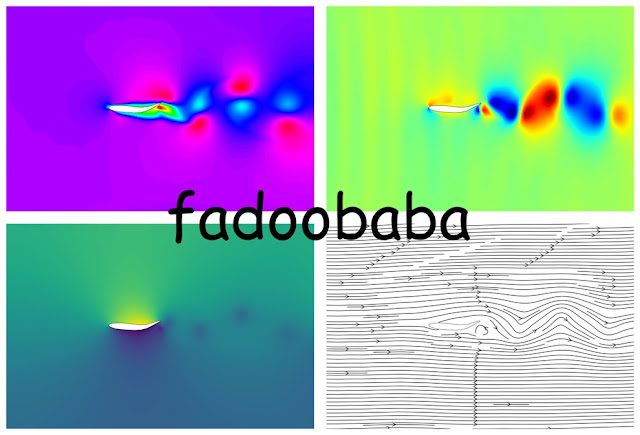
For the same grid and for solving the same problem, the time-step supported by equation 2 is 40x more as compared to equation 1. Therefore, the resultant compute per watt is 40x less for equation 2 as compared to equation 1 🤯. The code for equation 1 is shown first, followed by the code for equation 2. The results are compared via streamlines and pressure contours, with in Fig. 1. The benchmark case solved is of the lid-driven cavity which offer simple implementation and very complex flow physics. For validation of the code, refer to here, here and here.
Copyright <2025> <FAHAD BUTT>
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
If you are mad 👨🔬 enough to use this code in your scholarly 👨🏫 work, then please remember to cite: Fahad Butt (2025). S-PPE (https://fluiddynamicscomputer.blogspot.com/2025/11/a-simple-poissons-equation-for-pressure.html), Blogger. Retrieved Month Date, Year
MANDATORY NOTE: This method requires a GPU, if dear readers don't have a GPU then please stop being peasants... 🙉
Code 01
#%% import libraries
import cupy as cp
import matplotlib.pyplot as plt
#%% define parameters
l_cr = 1 # characteristic length
h = l_cr / 100 # grid spacing
dt = 0.00005 # time step
L = 1 # domain length
D = 1 # domain depth
Nx = round(L / h) + 1 # grid points in x-axis
Ny = round(D / h) + 1 # grid points in y-axis
nu = 1 / 400 # kinematic viscosity
Uinf = 1 # free stream velocity / inlet velocity / lid velocity
cfl = dt * Uinf / h # cfl number
travel = 10 # times the disturbance travels entire length of computational domain
TT = travel * L / Uinf # total time
ns = int(TT / dt) # number of time steps
Re = round(l_cr * Uinf / nu) # Osborne Reynolds and his number :)
#%% intialization (t = 0)
u = cp.zeros((Nx, Ny)) # x-velocity
v = cp.zeros((Nx, Ny)) # y-velocity
p = cp.zeros((Nx, Ny)) # pressure
X, Y = cp.meshgrid(cp.linspace(0, L, Nx), cp.linspace(0, D, Ny), indexing = 'ij') # spatial grid
#%% pre calculate for speed
P1 = h / (8 * dt)
P2 = (2 / Re) * dt / h**2
P3 = dt / h
P4 = 1 - (4 * P2)
#%% solve 2D incompressible Navier-Stokes equations
for _ in range(ns):
pn = p.copy()
p[1:-1, 1:-1] = 0.25 * (pn[2:, 1:-1] + pn[:-2, 1:-1] + pn[1:-1, 2:] + pn[1:-1, :-2]) - P1 * ((u[2:, 1:-1] - u[:-2, 1:-1] + v[1:-1, 2:] - v[1:-1, :-2]) - (u[2:, 1:-1] - u[:-2, 1:-1])**2 - (v[1:-1, 2:] - v[1:-1, :-2])**2 - (2 * (u[2:, 1:-1] - u[:-2, 1:-1]) * (v[1:-1, 2:] - v[1:-1, :-2]))) # pressure
p[0, :] = p[1, :] # dp/dx = 0 at x = 0
p[-1, :] = p[-2, :] # dp/dx = 0 at x = L
p[:, 0] = p[:, 1] # dp/dy = 0 at y = 0
p[:, -1] = p[:, -2] # dp/dy = 0 at y = D
un = u.copy()
vn = v.copy()
u[1:-1, 1:-1] = un[1:-1, 1:-1] * P4 - P3 * (un[1:-1, 1:-1] * (un[2:, 1:-1] - un[:-2, 1:-1]) + vn[1:-1, 1:-1] * (un[1:-1, 2:] - un[1:-1, :-2]) + p[2:, 1:-1] - p[:-2, 1:-1]) + P2 * (un[2:, 1:-1] + un[:-2, 1:-1] + un[1:-1, 2:] + un[1:-1, :-2]) # x momentum
u[0, :] = 0 # u = Uinf at x = 0
u[-1, :] = 0 # u = 0 at x = L
u[:, 0] = 0 # u = 0 at y = 0
u[:, -1] = Uinf # u = Uinf at y = D
v[1:-1, 1:-1] = vn[1:-1, 1:-1] * P4 - P3 * (un[1:-1, 1:-1] * (vn[2:, 1:-1] - vn[:-2, 1:-1]) + vn[1:-1, 1:-1] * (vn[1:-1, 2:] - vn[1:-1, :-2]) + p[1:-1, 2:] - p[1:-1, :-2]) + P2 * (vn[2:, 1:-1] + vn[:-2, 1:-1] + vn[1:-1, 2:] + vn[1:-1, :-2]) # y momentum
v[0, :] = 0 # v = 0 at x = 0
v[-1, :] = 0 # v = 0 at x = L
v[:, 0] = 0 # v = 0 at y = 0
v[:, -1] = 0 # v = 0 at y = D
Code 2
#%% import libraries
import cupy as cp
import matplotlib.pyplot as plt
#%% define parameters
l_cr = 1 # characteristic length
h = l_cr / 100 # grid spacing
dt = 0.002 # time step
L = 1 # domain length
D = 1 # domain depth
Nx = round(L / h) + 1 # grid points in x-axis
Ny = round(D / h) + 1 # grid points in y-axis
nu = 1 / 400 # kinematic viscosity
Uinf = 1 # free stream velocity / inlet velocity / lid velocity
cfl = dt * Uinf / h # cfl number
travel = 10 # times the disturbance travels entire length of computational domain
TT = travel * L / Uinf # total time
ns = int(TT / dt) # number of time steps
Re = round(l_cr * Uinf / nu) # Osborne Reynolds and his number :)
#%% intialization (t = 0)
u = cp.zeros((Nx, Ny)) # x-velocity
v = cp.zeros((Nx, Ny)) # y-velocity
p = cp.zeros((Nx, Ny)) # pressure
X, Y = cp.meshgrid(cp.linspace(0, L, Nx), cp.linspace(0, D, Ny), indexing = 'ij') # spatial grid
#%% pre calculate for speed
P1 = h / (16 * dt)
P2 = (2 / Re) * dt / h**2
P3 = dt / h
P4 = 1 - (4 * P2)
#%% solve 2D incompressible Navier-Stokes equations
for _ in range(ns):
pn = p.copy()
p[1:-1, 1:-1] = 0.25 * (pn[2:, 1:-1] + pn[:-2, 1:-1] + pn[1:-1, 2:] + pn[1:-1, :-2]) - P1 * (u[2:, 1:-1] - u[:-2, 1:-1] + v[1:-1, 2:] - v[1:-1, :-2]) # mass
p[0, :] = p[1, :] # dp/dx = 0 at x = 0
p[-1, :] = p[-2, :] # dp/dx = 0 at x = L
p[:, 0] = p[:, 1] # dp/dy = 0 at y = 0
p[:, -1] = p[:, -2] # dp/dy = 0 at y = D
un = u.copy()
vn = v.copy()
u[1:-1, 1:-1] = un[1:-1, 1:-1] * P4 - P3 * (un[1:-1, 1:-1] * (un[2:, 1:-1] - un[:-2, 1:-1]) + vn[1:-1, 1:-1] * (un[1:-1, 2:] - un[1:-1, :-2]) + p[2:, 1:-1] - p[:-2, 1:-1]) + P2 * (un[2:, 1:-1] + un[:-2, 1:-1] + un[1:-1, 2:] + un[1:-1, :-2]) # x momentum
u[0, :] = 0 # u = Uinf at x = 0
u[-1, :] = 0 # u = 0 at x = L
u[:, 0] = 0 # u = 0 at y = 0
u[:, -1] = Uinf # u = Uinf at y = D
v[1:-1, 1:-1] = vn[1:-1, 1:-1] * P4 - P3 * (un[1:-1, 1:-1] * (vn[2:, 1:-1] - vn[:-2, 1:-1]) + vn[1:-1, 1:-1] * (vn[1:-1, 2:] - vn[1:-1, :-2]) + p[1:-1, 2:] - p[1:-1, :-2]) + P2 * (vn[2:, 1:-1] + vn[:-2, 1:-1] + vn[1:-1, 2:] + vn[1:-1, :-2]) # y momentum
v[0, :] = 0 # v = 0 at x = 0
v[-1, :] = 0 # v = 0 at x = L
v[:, 0] = 0 # v = 0 at y = 0
v[:, -1] = 0 # v = 0 at y = D
 |
| Fig. 1, pressure range for both cases is 0 (blue) till 1 (red). |
If you want to hire me as your next shining post-doc or collaborate in research, please reach out! Thank you for reading!